Image Reconstruction#
Image reconstruction in MRI means converting the raw signal collected in k-space into images. For a fully-sampled acquisition using Cartesian sampling (e.g. on a grid), this can be done with a discrete Fourier Transform. In anticipation of advanced reconstruction methods, image reconstruction for MRI is formulated as a linear system.
Learning Goals#
Describe how images are formed
Describe how MRI raw data is reconstructed into an image
Manipulate and analyze MRI data
Reconstruct an image from raw data
Sorting the MRI Data into k-space#
Recall that the MRI signal is proportional to the Fourier Transform of the net magnetization of our object, evaluated at the k-space location defined by the gradients:
Note that this includes a dimension for the readout, \(t\), as well as different readouts for each TR, denoted by the subscript for the \(i^\mathrm{th}\) TR.
The data acquired during the readout will be sorted into data based on the k-space trajectory. The following example shows the Cartesian k-space trajectory, which acquired data in a raster fashion in k-space:
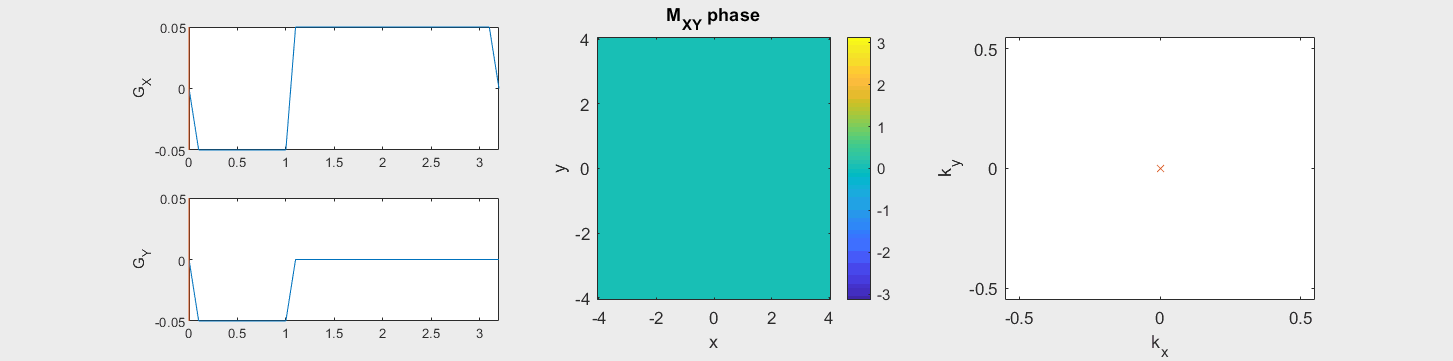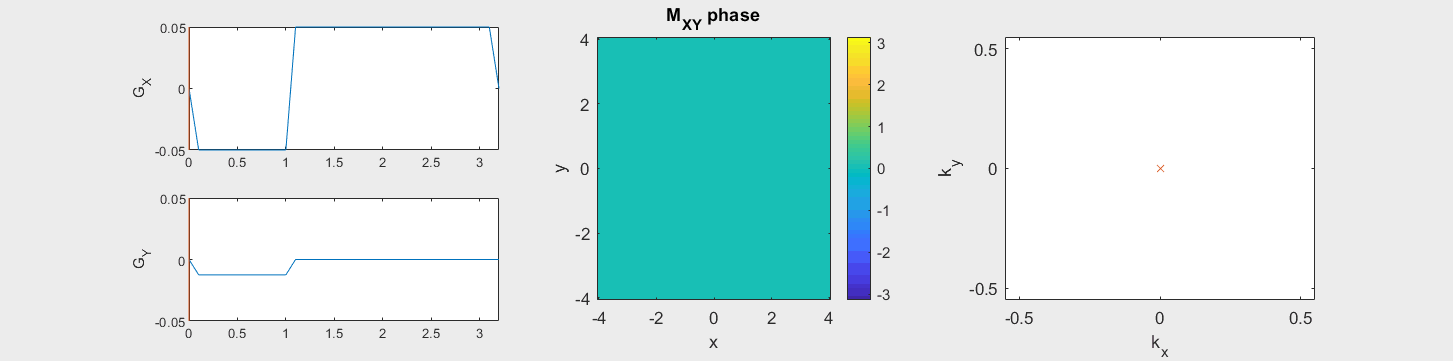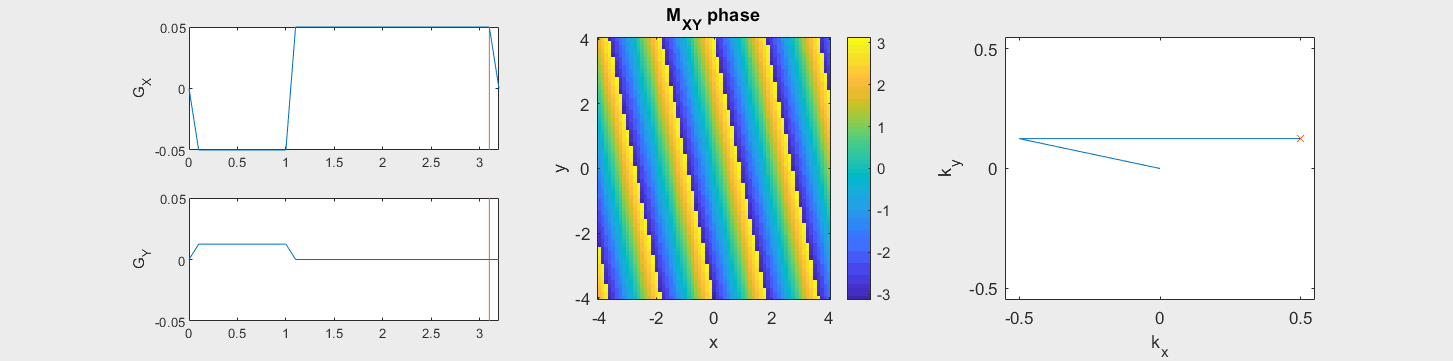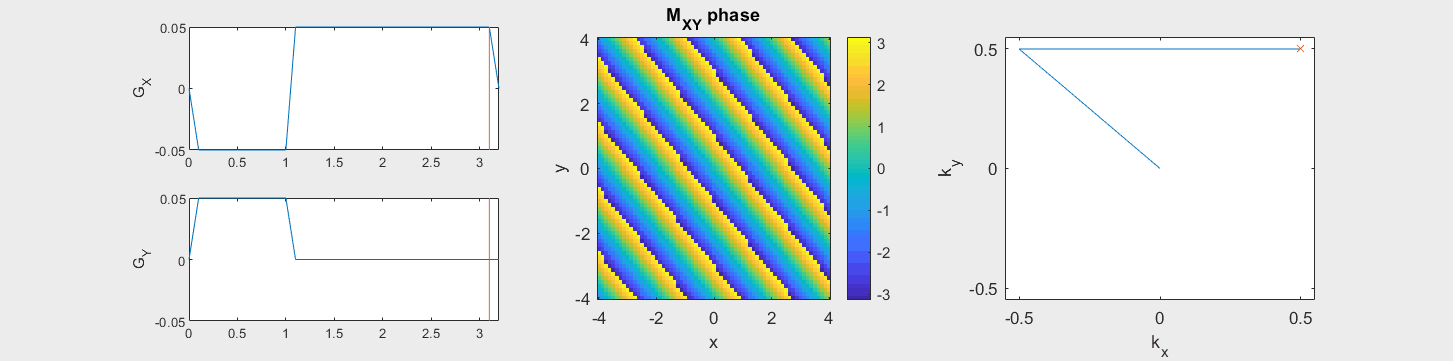
From this knowledge, the MR signal over time is stored in a data matrix with known k-space locations to create \(M(\vec{k})\).
Fourier Transform Reconstruction#
Once the data has been sorted into the corresponding lines in k-space, an inverse Fourier Transform is applied to reconstruct an image of the transverse magnetization
For typical Cartesian sampling, this can be done very simply with the 2D or 3D Fast Fourier Transform (FFT) algorithm.
The following animation illustrates typical k-space data as it would be acquired for different k-space lines (left) and the resulting image as more and more lines of k-space are accumulated

We can also acquire our k-space lines in a “center-out” or random ordering, shown below
Linear System Formulation#
Alternatively, the reconstruction can be formulated as a linear system, using a discrete Fourier Transform matrix, \(\mathbf{F}\). For this matrix, the entries are defined by k-space location being sampled, \(\vec{k}_i\) and the spatial locations, \(\vec{r}_j\):
In this case, the measurement is the discrete Fourier Transform of the image
and image reconstruction is performed by inverse discrete Fourier Transform, which for fully sampled 2D FT imaging is well defined by matrix inversion:
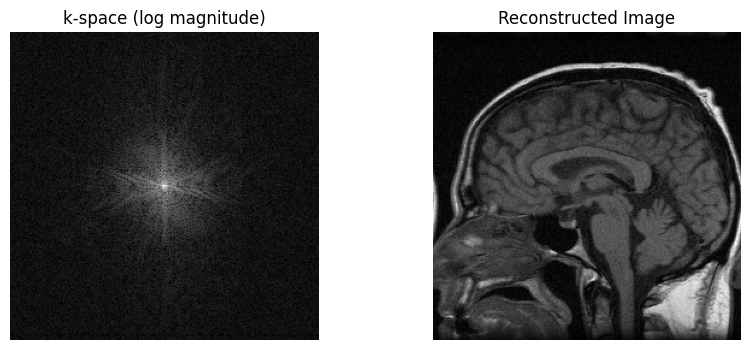
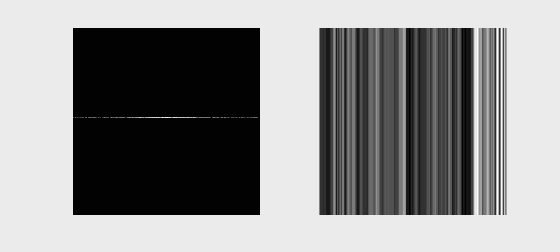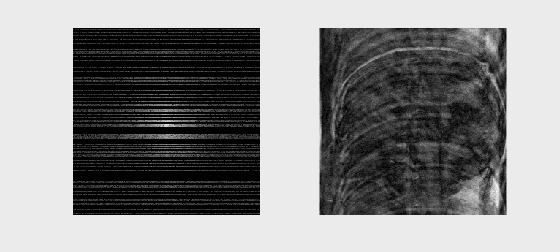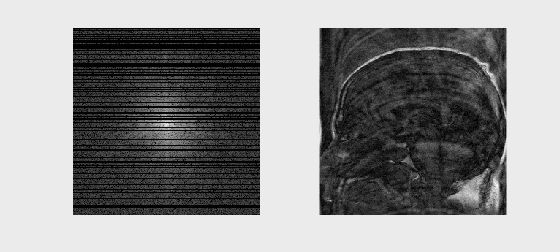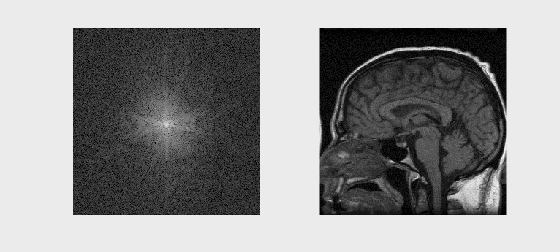
# Sample Image Reconstruction
import numpy as np
from scipy.io import loadmat
import matplotlib.pyplot as plt
# Load k-space data from .mat file
mat = loadmat('../Data/se_t1_sag_data.mat')
kspace = mat['data']
# Reconstruct the image by inverse FFT
recon_image = np.fft.fftshift(np.fft.ifft2(np.fft.ifftshift(kspace)))
# Plot k-space data and reconstructed images
fig, axs = plt.subplots(1, 2, figsize=(10, 4))
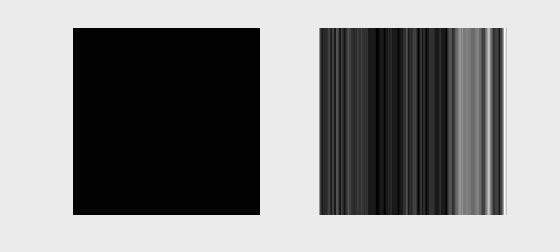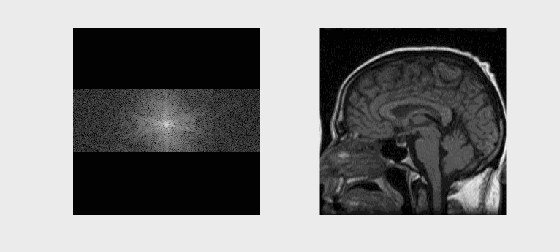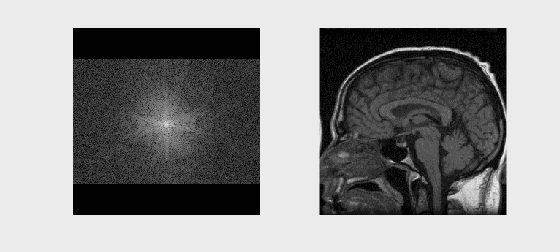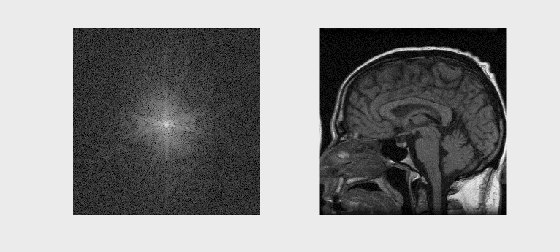
axs[0].imshow(np.log(np.abs(kspace) + 1e1), cmap='gray')
axs[0].set_title('k-space (log magnitude)')
axs[0].axis('off')
axs[1].imshow(np.abs(recon_image), cmap='gray')
axs[1].set_title('Reconstructed Image')
axs[1].axis('off')
(-0.5, 255.5, 255.5, -0.5)